PCA and LDA are two popular dimensionality reduction methods commonly used on data with too many input features. In many ways the two algorithms are similar, but at the same time very dissimilar. This article highlights some of the similarities and dissimilarities between these two popular algorithms.
Let’s remind ourself about how these two algorithms work.
Principal Component Analysis or PCA for short is a commonly used unsupervised linear transformation technique. PCA reduces the number of dimensions by finding the maximum variance in high dimensional data.
Linear Discriminant Analysis or LDA for short is a supervised method that takes class labels into account when reducing the number of dimensions. The goal of LDA is to find a feature subspace that best optimizes class separability.
Algorithm’s Learning mechanism
Both algorithms rely on decomposing matrices of eigenvalues and eigenvectors, but the biggest difference between the two is in the basic learning approach. Where PCA is unsupervised, LDA is supervised.
PCA reduces dimensions by looking at the correlation between different features. This is done by creating orthogonal axes – or principal components – with the direction of maximum variance as a new subspace.
Essentially PCA generates components based on the direction in the data where there is most variance – eg the data is most spread out. This component is referred to as both principals, and eigenvectors, and represent a subset of the data that contains most of the information – or variance – of our data. LDA on the other side does almost the same, but it contains a “pre-processing” step calculating mean vectors from the class labels before extracting the eigenvalues.
Step by step this will look like this
LDA
1 – For each class label: compute the d-dimensional mean vector
2 – Construct a scatter matrix within each class, and between each class.
This means that we first generate a mean vector for each label, so if we have three labels, we will generate three vectors.
Then with these three mean vectors, we generate a scatter matrix for each class, and then we sum up the three individual scatter matrices into one final matrix. We now have the within each class matrix.
To generate the between each class matrix we take the overall mean value from the original input dataset, and then subtract the overall mean with the mean of each mean vector as a dot product of itself. This is best explained with the equation given below, where m is the overall mean from the original input data.
$$ S_B = \sum\limits_{i=1}^{c} N_{i} (\pmb m_i – \pmb m) (\pmb m_i – \pmb m)^T $$
PCA
1 – Construct the covariance matrix by taking the joint covariance – or correlation in some cases – between each pair in the given vector.
Then with the generated matrix we..
1 – Compute eigenvectors and eigenvalues of the matrix
2 – Sort the eigenvalues by decreasing order to rank the eigenvectors
3 – Get the k eigenvectors which corresponds to the k largest eigenvalues
4 – Construct a projection Matrix from the top k eigenvectors
5 – Transform the original input dataset with the newly created projection
This was a comparison between PCA and LDA, and as we can see they are very similar; both are linear transformation techniques that decompose matrices of eigenvalues and eigenvectors. The main difference is that LDA takes into class labels into account, where PCA is unsupervised and does not.
Dictionary:
- Eigenvectors: Scaled version of the original vector
- Eigenvalues: Scalar used to transform an Eigenvector
Auto Amazon Links: No products found.


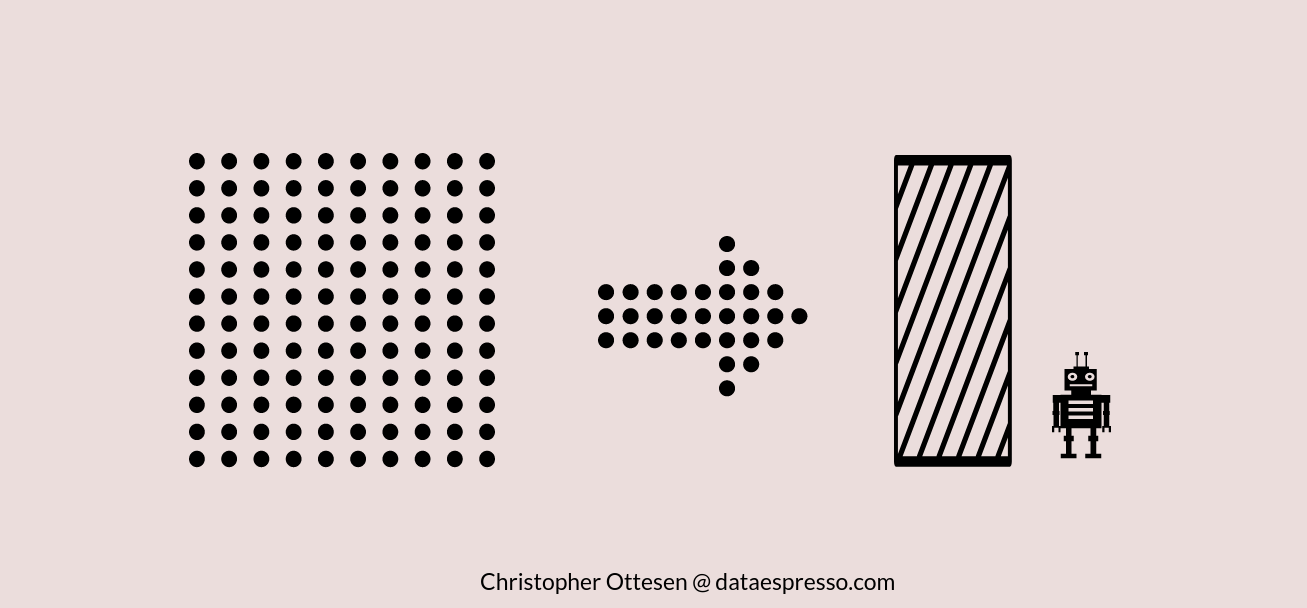







8 responses to “Comparison between PCA and LDA”
I am regular reader, how are you everybody? This piece of writing posted at this site is genuinely good.
You really make it seem so easy with your presentation however I to
find this matter to be really something that I think I might by no means understand.
It seems too complicated and very large for me. I am taking a look forward on your subsequent put up, I will
try to get the dangle of it!
I’ll immediately take hold of your rss feed as
I can not in finding your email subscription link or newsletter service.
Do you have any? Kindly permit me understand so that I could subscribe.
Thanks.
Hello! You can subscribe here: https://dataespresso.com/en/newsletter/
Thanks 🙂
I couldn?t refrain from commenting. Perfectly written!|
Good day very cool blog!! Guy .. Beautiful .. Wonderful .. I will bookmark your blog and take the feeds also?I’m satisfied to seek out a lot of helpful info right here in the publish, we want work out extra strategies on this regard, thank you for sharing. . . . . .
I don?t even know how I ended up here, but I thought this post was great. I don’t know who you are but certainly you are going to a famous blogger if you are not already 😉 Cheers!
fantastic post, very informative. I wonder why the other specialists of this sector don’t notice this. You must continue your writing. I am sure, you have a great readers’ base already!